2012/13 flu update: We're re-running the primer on flu biology (reproduced in full below) that we developed for the 2009 pandemic in order to keep you up-to-date with what's turning out to be a busy flu season. The fraction of people visiting physicians for flu-like symptoms rose from 2.8 percent to 5.6 percent during the last few weeks of 2012, according to statistics from the CDC. Since then, it seems to have started to decline.
At its peak, while this year was below some recent years (in the 2009 pandemic, that rate was 7.7 percent), that number still represents a lot of sick people. And, unfortunately, it represents fatalities. So far, the CDC has been made aware of 40 pediatric deaths from influenza this season, and that number is likely to rise over the coming weeks. Rates of hospitalization peaked at about 8.1 per 100,000 people.
The majority of people seem to be infected with the influenza A species of flu virus (although some influenza B is also circulating), predominantly H3N2—for more on that nomenclature, see below. The good news for those who haven't been infected yet is the majority of the viruses the CDC has seen are covered by this year's vaccine. So, if you got a shot, chances are good that you'll be protected.
Our original coverage follows:
Swine flu, bird flu, H1N1—tracking the influenza virus can be a confusing task, not generally made easier by the fact most people only attempt to do so when addled by flu symptoms or in the midst of worries about a potential pandemic. We recognize the latter appears to apply to the current situation, but we'll do our part to try to explain a bit of the biology of the virus. Putting together this explanation was made a bit challenging by the fact that anyone we could find who has detailed knowledge of the influenza virus appears to be busy actually working on the current outbreak.
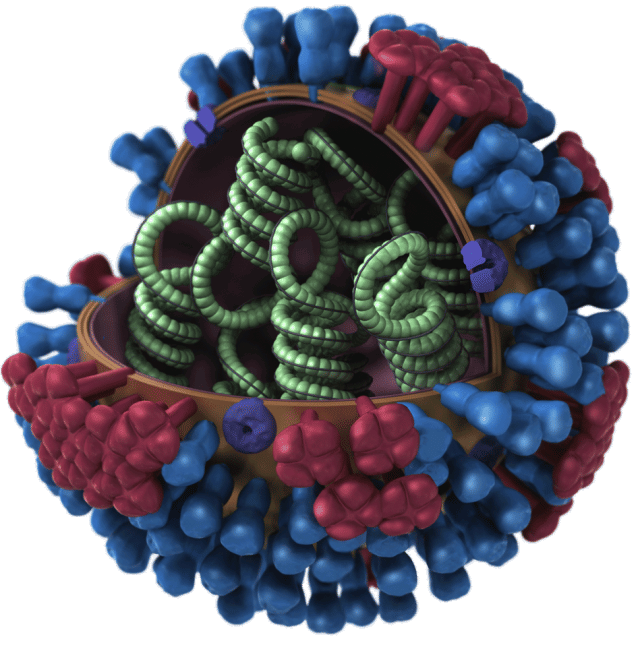
On the surface: the HxNx nomenclature
Like most viruses, a spreading swine flu virus has a coat formed of proteins which surround the genetic material that allows the virus to hijack a cell and reproduce. These coat proteins are critical in a variety of ways: they determine which cells the virus can latch onto and infect and, being exposed, they're the things antibodies recognize when your body generates an immune response to the virus. For the flu virus, the major coat proteins are called hemagglutinin and neuraminidase—the H and N of the commonly used nomenclature for identifying these viruses.
Given the large and diverse flu virus population, there are many variants of each of these two proteins, but they fall roughly into a limited number of classes: according to the CDC, there are 16 known hemagglutinin classes, and nine of the neuraminidase. As such, it's possible to identify categories of viruses based on their specific combination of these two genes, leading to the typical nomenclature that we see in press reports about influenza: H1N1, H3N2, etc.
It would be reassuring if these broad categories lined up nicely with the virus' hosts, so that this nomenclature was informative. Unfortunately, given the influenza virus' tendency to mutate and hop among species, that's not the case. So, for example, pigs carrying the virus are most often infected with an H1N1 subtype but, according to the World Health Organization, at least three other subtypes have also been found. The viruses currently known to circulate in humans are H1N1, H1N3, and H3N2. Two of those also appear in pigs, and the CDC indicates that all known subtypes of influenza virus can circulate in bird populations.
To add to the confusion, there is a lot of variability within these broad groupings. That's why, even if you've received a vaccine against an H1N1 virus, it may not protect you against the H1N1 virus that actually starts circulating in a given year. The differences within a group can also contribute to the host range, virulence, and other properties of the virus. So, even if a given subtype of virus doesn't circulate in humans at the moment, there's no guarantee that it won't at some point in the future.
Beyond the surface
Although these two genes are rather important, a quick look at the completed influenza genomes reveals there's a lot more going on in a typical flu virus. Two other genes help hold the surface coat together, and a third packages up the virus's genome, both inside the cell and out. Three other proteins manipulate the host, shutting down host defenses, getting the virus' genome made into proteins, and killing the cell when it's time to spread.
Finally, the influenza virus' genome exists as RNA, rather than DNA, and the cell's enzymes are not prepared to duplicate that or transcribe it into the messenger RNAs that get made into proteins. So, the flu virus brings its own, with three genes encoding RNA-dependent polymerases.
How important are the rest of these proteins? A study examined the mutations required for an influenza virus to adapt to growing in a new host; it found 14 of them, scattered widely through the viral genome. It's possible to make a rational case that changes in any one of these genes could dramatically change the dynamics of an infection, and thus (potentially) its lethality.
Rearrangements, hosts, and reservoirs
That said, examining the genome of the virus as a whole can allow some general classifications. Collections of sequence differences are often commonly found in viruses specific to a given host, hence the general classification of things like swine flu and bird flu. But again, these are only rough guidelines—swine flu may be typically found in pigs, but it hops into humans on a regular enough basis to be a common health concern. In fact, the ability of a flu virus to lurk in other species for a while before returning to plague humans is one of the reasons we often refer to those species as reservoirs (presumably, pigs would view us as a reservoir for their flu pandemics if they had the mental wherewithal). Essentially, the flu virus treats most tetrapods as a giant ecosystem with a variety of niches that it can adapt to.
The series of reservoirs that the virus can move between creates two possibilities for new viral properties to evolve. The first is that a single mutation may have very different effects in different species. A change that slows the virus down, allowing it to evade immune detection in, say, pigs, could allow it to progress rapidly in human populations.
The other issue is that a single cell can wind up harboring copies of viruses with diverse origins, which can then undergo rearrangements. Different viral genes reside on different segments of RNA, and these can wind up mixing fairly easily. Experimental evidence also indicates that some sort of rearrangement is possible within a segment: put two viruses, each with a different gene inactivated, into a cell, and a single, healthy virus can emerge.
This sort of mixing of genetic material between viruses doesn't appear to be common, but there is evidence that rearrangements that mix pieces of different viruses have happened multiple times in recent history, and some of these have been responsible for near pandemics. That's not to say that any rearranged virus is bad—a lot of them probably generate a harmless virus, so we never see them—but it does seem that the process is more likely to create something new, at least from our immune system's perspective.
All of which brings us back to the current virus. The CDC's Anne Schuchat (an MD and Rear Admiral), who directs the National Center for Immunization and Respiratory Diseases, describes the virus as follows: "We know so far that the viruses contain genetic pieces from four different virus sources. This is unusual. The first is our North American swine influenza viruses. North American avian influenza viruses, human influenza viruses and swine influenza viruses found in Asia and Europe." If it's being called swine flu, that's apparently because two out of four parts of it are from there, but it clearly has a complicated evolutionary history.
Again, this doesn't mean that the virus is especially threatening, but it does suggest that it will be harder to base our expectations of its behavior on past flu outbreaks. And that's probably why health authorities are so anxious to monitor its spread and get some hard data on how it behaves and the specific symptoms it causes.
reader comments
68